Evaluate Effectiveness, Specificity and Safety Evaluation of New Potential Drug Targets
OmniBrowser provides various collection across cancer type, cell type and differential gene expression bar plots. User can get the target’s cancer type specific expression according to this information, then validate the effectiveness and the specificity a potential drug target.
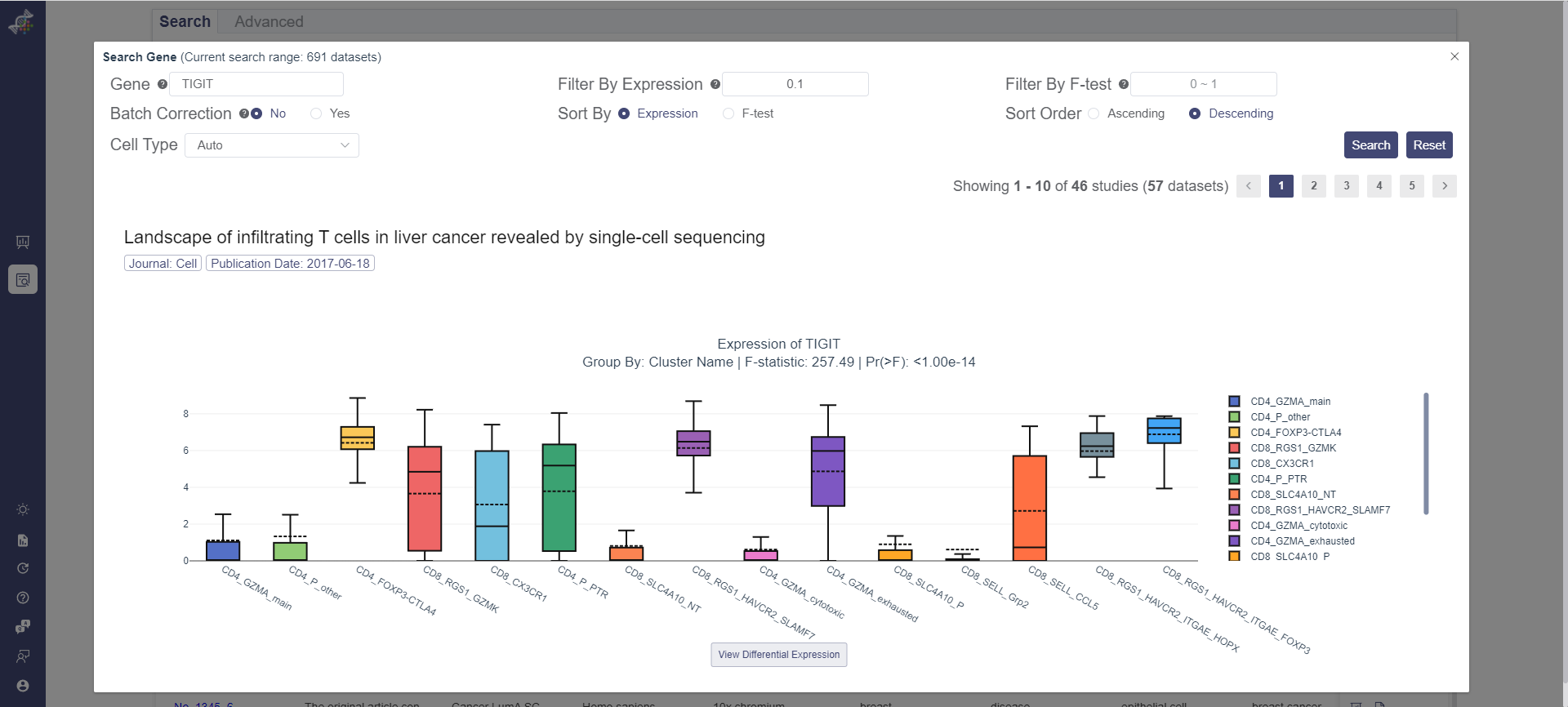 Search the expression of TIGIT
Search the expression of TIGIT
For instance, TIGIT is a well-known drug target. You can use OmniBrowser to search TIGIT through all human cancer datasets (341), and find out 65 TIGIT expressed datasets. In these datasets, TIGIT generally mostly expressed in T cell, NK cell. In some specific cancer type, it also expressed in ILCs. And the dataset relates to many cancer types such as liver cancer, colon cancer, renal cancer, breast cancer, lung cancer, melanoma, head and neck cancer, acute leukemia, glioma. This information can tell us which cancer types that TIGIT may be effective and the specificity of it.
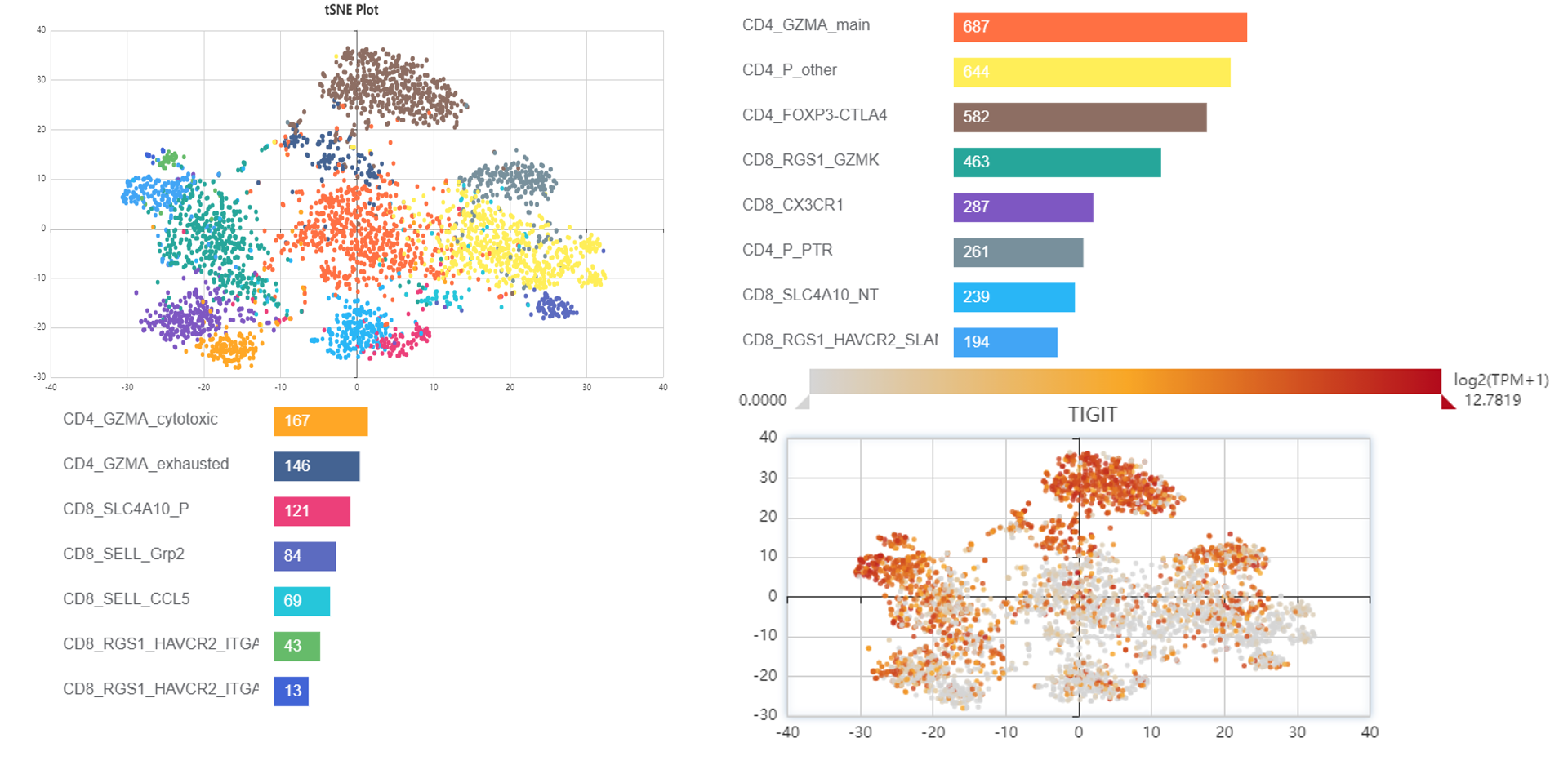 TIGIT expression cell types
TIGIT expression cell types
For further exploring, you can select a dataset, such as (No_13_1)“Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing”. We can see the subcluster expression level of TIGIT in this data.
As you can see, TIGIT expressed in:
CD4+ T cell (Helper T cell):
peripheral Tregs;
CD4-CTLA4: tumor Tregs
CD4-CXCL13: exhausted CD4+ T cells
CD8+ T cell (Cytotoxic T cell)
CD8-LAYN: exhausted CD8+ T cells
CD8-CX3CR1: effector memory CD8+ T cells
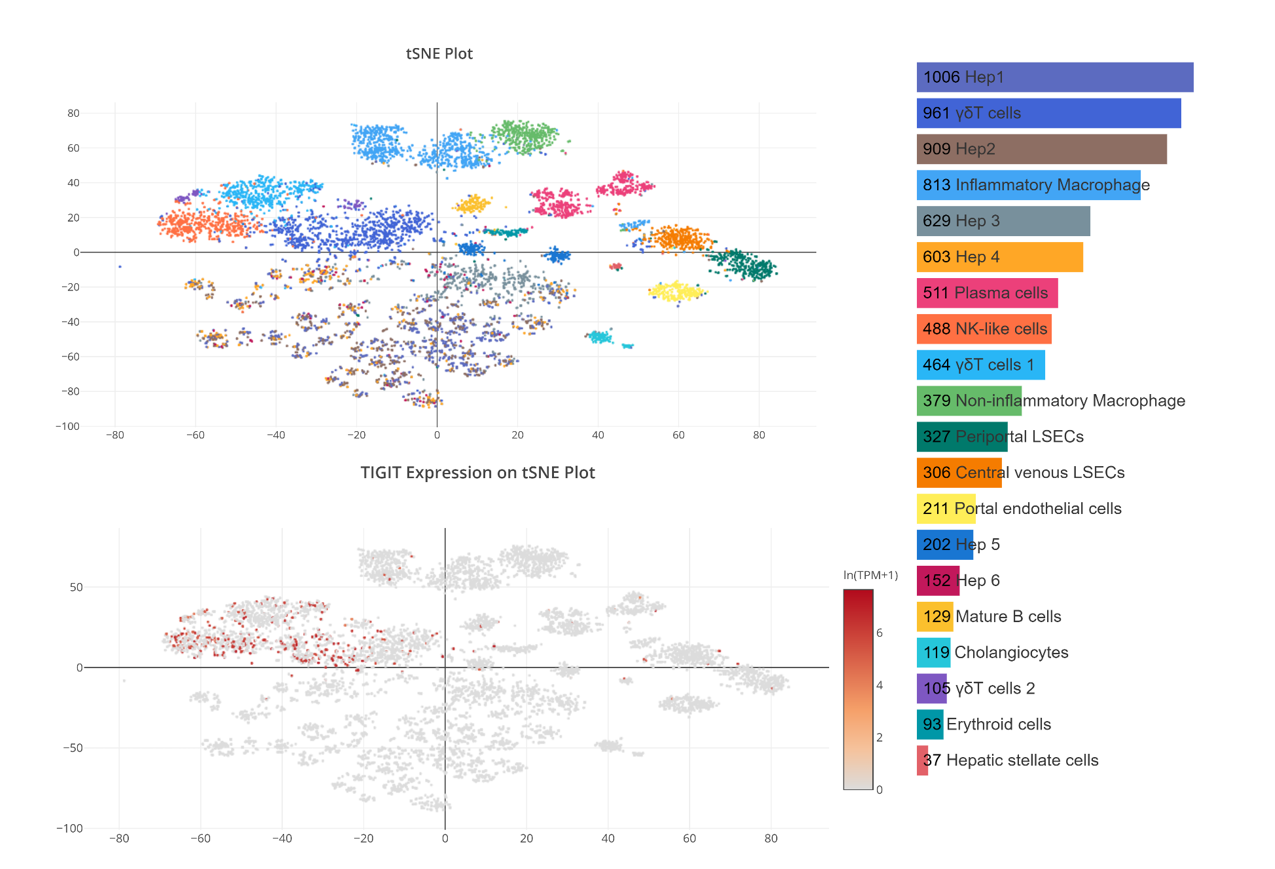 Hepatotoxicity Evaluation of TIGIT
Hepatotoxicity Evaluation of TIGIT
On the other hand, OmniBrowser has the collection of normal cells across varies of tissues, which can help us evaluate the safety of the target. For instance, we can do hepatotoxicity evaluation of TIGIT. After filtering the data by species, tissues and research topic, you can search genes and select the data which has hepatocytes. Then you can see that TIGIT are specific expressed in NK cells and T cells and not expressed in hepatocytes cells. Therefore, we can know that TIGIT has no Hepatotoxicity.